All published articles of this journal are available on ScienceDirect.
Cross-Clade Recognition of HIV-1 CAp24 by CD4+ T Cells in HIV-1-Infected Individuals in Burkina Faso and Germany
Abstract
The presence of antigen-specific cellular immune responses may be an indicator of long-term asymptomatic HIV-1-disease. The detection of cellular immune responses to infection with different subtypes of HIV-1 may be hampered by genetic differences of immunodominant antigens such as the capsid protein CAp24. In Nouna, Burkina Faso, HIV-1 circulating recombinant forms CRF02_AG and CRF06_cpx are the 2 major strains detectable in HIV-1-infected individuals, while subtype B strains prevail in Europe and North America. Amino acid sequences of CAp24 were assessed in blood samples from 10 HIV-1-infected patients in Nouna, Burkina Faso. Production of interferon-gamma (IFN-γ) in peripheral blood CD4+ lymphocytes in response to recombinant HIV-1 proteins derived from clade B (including CAp24NL4-3) was measured using a modified flow-cytometry-based whole blood short term activation assay (FASTimmune, BDBiosciences). IFN-γ production following stimulation with a whole length CAp24 protein derived from clade B (CAp24NL4-3) was additionally quantified in comparison to a CAp24 protein derived from CRF02_AG (CAp24BD6-15) in 16 HIV-1-infected patients in Heidelberg, Germany. Amino acid sequence identity of CAp24 obtained from patients in Nouna ranged between 86 and 89% when compared to the clade B CAp24NL4-3 consensus sequence, between 90 and 95% when compared to the circulating recombinant form CRF06_CPX consensus sequence, and between 92 and 96% when compared to the CAp24BD6-15 consensus sequence. Significant numbers of HIV-1-specific CD4+ lymphocytes producing IFN-γ were detected in 4 of 10 HIV-1-infected patients. In 7 of 16 patients in Heidelberg, recombinant CAp24BD6-15 stimulated IFN-γ-production in CD4+ lymphocytes to a similar extent as the clade B-derived CAp24NL4-3. Thus, antigen-specific CD4+ lymphocytes from both West African and European patients infected with different strains of HIV-1 show relevant cross-clade recognition of HIV-1 CAp24 in a flow-cytometry-based whole blood short term activation assay.
INTRODUCTION
The presence of antigen-specific cellular immune responses may be an indicator of long-term asymptomatic HIV-1-disease [1]. However, the detection of cellular immune responses to infection with different subtypes of HIV-1 may be hampered by strain-specific differences of immunodominant antigens especially in the CD4+ subset. Norris et al. [2] observed that many of the naturally occurring HIV-1 Gag epitope variants from clades A, B, C, D, and AE induced less than 50% of the interferon-γ (IFN-γ)-response elicited by antigens based on the clade B consensus sequence in CD4+ T cell clones from individuals with vigorous HIV-1-specific T helper cell activity. On the other side, cross-reactive cytotoxic T lymphocytes against antigens derived from clade A, B, C, or D were observed in patients infected with different HIV-1 subtypes in Uganda [3] and HIV-1 Gag peptide pools were able to elicit broad reactivity of CD8+ T cells across several subtypes of HIV-1 in chronically infected patients in Malawi and the US [4] or the Netherlands [5].
MATERIALS AND METHODS
The Nouna Health Research Center (Centre de la Réchèrche en Santé de Nouna, CRSN) and the Department of Virology of the University of Heidelberg, Germany, conduct a longitudinal trial of prevention of mother-to-child transmission (PMTCT) of the human immunodeficiency virus type-1 (HIV-1) in Nouna, north-western Burkina Faso (West-Africa) [6]. The study was approved by the Institutional Ethics Committees at the CRSN and the University of Heidelberg. Initial molecular analysis of blood samples from the research area identified the circulating recombinant forms CRF02_AG and CRF06_cpx as the predominant strains in HIV-1-infected individuals in Nouna [7].
Functional Analysis of HIV-1-Specific T Helper Cells
In a field-study in Nouna we screened 24 HIV-1-seropositive (HIV-1+) individuals (4 males and 20 females) for HIV-1-specific cytokine responses in peripheral blood CD4+ lymphocytes. Their median age was 33 years (range, 18 to 62). Eight patients received antiretroviral therapy with a combination of Zidovudine, Lamivudine, and Nevirapine at the time of the study. Plasma viral load levels were determined in 20 patients and ranged from below detection threshold (<50 copies per ml) to 2.2*106 copies per ml (median 3.1*105 copies per ml). CD4 counts ranged from 87 to 914 cells per µl (median 390 cells per µl).
All experiments in Nouna were performed on a FACScanTM flow cytometer equipped with CellQuestTM pro software for data acquisition and analysis. Antigen-specific CD4+ lymphocytes were quantified in freshly drawn heparinised peripheral venous blood by flow cytometry using FASTimmuneTM reagents and protocols obtained from BD Biosciences (Erembodegem, Belgium, EU). T cells were stimulated for 9 hours at 37°C using anti-CD28/anti-CD49d mAbs (1 µg/ml) either alone or in combination with either Staphylococcal Enterotoxin B (SEB, 5 µg/ml; Sigma-Aldrich, München, Germany), or a mixture of recombinant proteins derived from HIV-1 clade B, including the capsid protein CAp24NL4-3 (25 µg/ml as endotoxin-free solution in phosphate-buffered saline) [8], the HIV-1 regulator protein Tat ARP682.2 (8 µg/ml), and the HIV-1 regulator Rev ARP663.2 (12.5 µg/ml), the latter two obtained through NIBSC Centralised Facility for AIDS Reagents (EU Programme EVA AVIP, UK medical research council). During the last 4 hours of stimulation 10 µg/ml Brefeldin A were added.
CD4+ lymphocytes were identified by a combined SSC/FL-3 and FSC/SSC gating strategy and the proportion of CD69+/INF-γ+ double-positive cells was expressed as percentage of CD4+ lymphocytes producing IFN-γ. The statistical significance of the difference between the percentage of IFN-γ-producing CD4+ lymphocytes after antigen stimulation and the corresponding unstimulated control was tested using the following power calculation: N = [2 * pav * (1*- pav) *10.5] / Δ², where N is the number of events in each sample needed for a statistical significance of p<0.05 with a power of 90%, pav is the average proportion between the background and test samples, and Δ is the difference between these two proportions [9].
Amino Acid Sequence Analysis of CAp24
Four spots of EDTA buffy coat from each patient were dried at 37°C on IsoCode Card and DNA was extracted later according to the manufacturer’s recommendations (Schleic-her und Schuell, Dassel, Germany). DNA was used as template for nested PCR with the following combinations of oligonucleotides (each 20 µM; Invitrogen, Groningen, The Netherlands): forward primers CA1:1056-1088 (TCA AAG GAT AGG TAA AAG ACA CCA AGG AAG C), CA3:1109-1139 (TAC AAA ATA AGA GCA AAA AGA CAC AGC A), GAG1:789-815 (TAT GGG TGC GAG AGC GTC GGT ATT AAG), and GAG3:836-875 (TGG GAA AAA TTC GGT TAA GGC C); reverse primers CA2:1169-1198 (TCT GGC TAG GTG TCC TTT GCC ACA GTT GAA), and CA4:1920-1952 (TTT TTG GCC CCT AAA ATT GCT TTT CTG CAT). A standard reaction of 50 µl contained 2 µl template, 41 µl dH20, 0.5 µl Taq-polymerase, 0.5 µl forward primer, 0.5 µl reverse primer, and 0.5 µl dNTP (dATP, dGTP, dCTP, dTTP). It was performed in 40 cycles (30 sec at 94°C, 45 sec at 45°C, 1 min at 72°C) after initial denaturation for 2 min at 94°C and a final extension of 5 min at 72°C. DNA fragments were cleaned with Nucleo Spin Extract kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s recommendations.
Sequencing was done by the primer walking method using the dye terminator cycle sequencing kit on a CEQ 2000 capillary sequencer (Beckman Coulter, CA, USA). Contigs were assembled using the Contig Assembler of the Vector NTI Suite (Infomax, Oxford, UK). Sequences obtained were aligned with HIV-1 subtype reference sequences using the Seqpublisher HIV database (www.hiv-web.lanl.gov).
Cross-Clade Recognition of CAp24 by T Helper Cells
We then purified both clade B-derived recombinant CAp24NL4-3 and CRF06_AG-derived CAp24BD6-15 [10] and compared the ability of the full length proteins to elicit HIV-1-specific cytokine production at a concentration of 20 µg/ml in peripheral blood CD4+ lymphocytes of 16 HIV-infected patients in Heidelberg, Germany. All patients had CD4 counts above 200 cells per µl and plasma viral load levels of at least 2.0*103 copies per ml. Patients were infected either with clade B (n = 12), clade A (n = 2), clade C (n = 1), or CRF01_AE (n = 1). The experiments were performed on a FACScalibur( flow cytometer with a slightly modified activation assay using CD154+/IFN-γ+ instead of CD69+/INF-γ+ double-positive CD4+ cells for quantification of antigen-specific cytokine responses [11].
RESULTS AND DISCUSSION
Flow-cytometry based functional screening revealed significant IFN-γ-responses in CD4+ lymphocytes of 8 out of 24 patients. Two of 7 responded to CAp24NL4-3 alone, 2 of 4 to CAp24NL4-3 and Tat, and 4 of 13 to CAp24NL4-3, Tat, and Rev. No correlation between the percentage of IFN-γ-producing HIV-1 specific CD4+ lymphocytes and either the CD4+ T cell counts or the plasma viral load levels was observed.
It has been shown that the capsid protein CAp24 is among the most conserved immunodominant HIV proteins [12]. We therefore specifically assessed amino acid sequences of CAp24 in blood samples from 10 patients in Nouna (6 untreated, 4 receiving antiretroviral combination therapy) in order to calculate sequence identity between patient samples and consensus sequences of clade B and circulating recombinant forms predominant in Nouna.
The resulting amino acid sequences are shown in Table 1. Amino acid sequence identity of capsid proteins ranged between 86 and 89% when compared to the clade B CA_NL4-3 consensus sequence, between 90 and 95% when compared to the circulating recombinant form CRF06_cpx consensus sequence, and between 92 and 96% when compared to the CRF02_AG consensus sequence previously determined in our laboratory [7]. CD4+lymphocytes from 3 untreated patients with CD4 counts above 200 cells per µl and plasma viral load levels above 105 copies per ml had significant IFN-γ-responses upon stimulation with the clade B-based antigen preparations described above, but no specific amino acid positions that may determine HIV-1-specific CD4+ lymphocyte responses could be identified.
Alignment of Amino Acid Sequences of CAp24 Retrieved from Plasma Viral RNA of 10 HIV-1-Infected Patients in Nouna Compared to the Clade B Laboratory Strain NL4-3 and the Circulating Recombinant Forms CRF06_cpx and CRF02_AG.
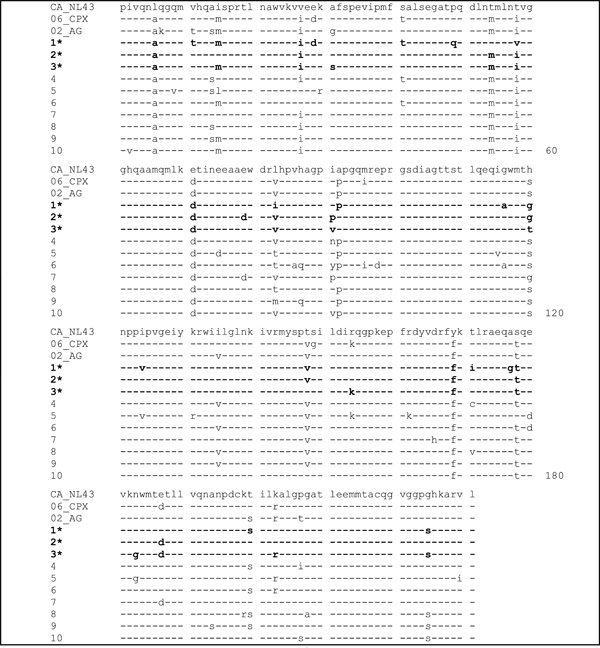 |
The asterisks
(*) and lines in bold indicate patients with a significant IFN-γ response of CD4+ lymphocytes after in vitro stimulation with recombinant CAp24NL4-3, dashes indicate (-) amino acid identity to the clade B laboratory strain NL4-3.
When the ability of the full length proteins CAp24NL4-3 and CAp24BD6-15 to elicit HIV-1-specific cytokine production in peripheral blood CD4+ lymphocytes was directly compared in 16 HIV-infected patients in Heidelberg, similar frequencies of HIV-1-specific CD4+ lymphocytes were detected (Fig. 1). Both antigens gave concordant results in 13 patients (Pearson's r = 0.91; p<0.001) revealing significant IFN-γ-production in 7 patients (3 on antiretroviral combination therapy, 4 untreated) and no significant activation in 6 patients (5 on antiretroviral combination therapy, one untreated). Responders were infected either with clade B (n = 5), clade A (n = 1), or CRF01_AE (n = 1). Non-responders were infected with clade B (n = 5), or clade C (n = 1). Discordant results were seen in 3 patients (one on antiretroviral combination therapy, 2 untreated, all infected with clade B).
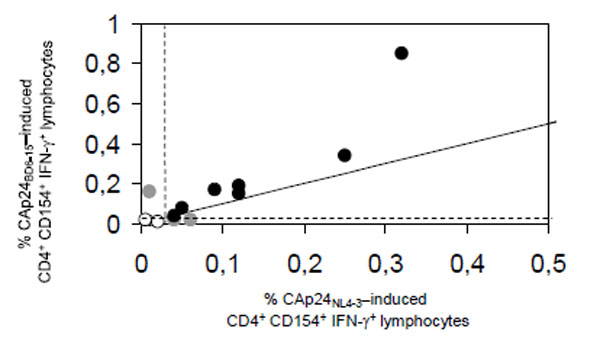
Correlation of antigen-specific CD4+lymphocyte responses elicited by HIV-1 clade B-derived CAp24NL4-3 (x-axis) and HIV-1 CRF02.AG-derived CAp24BD6-15 (y-axis); Pearson‘s r = 0.91, p<0.001). The solid line indicates the identity line, broken lines indicate the threshold for statistically significant production of interferon-γ (IFN-γ). Black dots indicate patients with concordant, grey dots patients with discordant, and circles patients without significant production of IFN-γ.
Of interest, CAp24BD6-15 yielded slightly higher percentages of cytokine-producing CD4+ lymphocytes in nearly all patients studied (Fig. 1). Furthermore, at both study sites we did not observe unspecific stimulation of these cells in blood samples from healthy adult laboratory personnel by neither CAp24NL4-3 nor CAp24BD6-15.
We conclude from our data that there is significant cross-clade recognition of the full-length proteins CAp24NL4-3 and CAp24BD6-15 by HIV-1 specific CD4+ lymphocytes in patients in Europe and in Burkina Faso who are infected with different strains of HIV-1. The study showed that FACS-based testing of CD4+ T cell function is feasible in a rural West African research area – even if it may be difficult to reproduce the conditions needed for high-throughput testing of HIV-1-specific CD4+ T cells [1] outside a major urban research center.
ACKNOWLEDGEMENTS
The study was supported by a research grant from Deutsche Forschungsgemeinschaft (SFB 544, project A6). We are indebted to clinical staff and patients in Nouna and in Heidelberg for their willingness to participate in the study. We gratefully acknowledge the help of Adama Compaoré and Nestor S. Dembélé with flow cytometry in Nouna. Thanks are due to Hans-Peter Spengler and Frans Nauwelaers, BD Biosciences,for their continuous interest and support, and to Lassana Sangaré and the staff at CHUYO, Ouagadougou, for determination of plasma viral load levels.


